You need to agree to share your contact information to access this model
This repository is publicly accessible, but you have to accept the conditions to access its files and content.
- This model and associated code are released under the CC-BY-NC-ND 4.0 license and may only be used for non-commercial, academic research purposes with proper attribution.
- Any commercial use, sale, or other monetization of the HistoPLUS model and its derivatives, which include models trained on outputs from the HistoPLUS model or datasets created from the HistoPLUS model, is prohibited and requires prior approval.
- By downloading the model, you attest that all information (affiliation, research use) is correct and up-to-date. Downloading the model requires prior registration on Hugging Face and agreeing to the terms of use. By downloading this model, you agree not to distribute, publish or reproduce a copy of the model. If another user within your organization wishes to use the HistoPLUS model, they must register as an individual user and agree to comply with the terms of use. Users may not attempt to re-identify the deidentified data used to develop the underlying model.
- This model is provided “as-is” without warranties of any kind, express or implied. This model has not been reviewed, certified, or approved by any regulatory body, including but not limited to the FDA (U.S.), EMA (Europe), MHRA (UK), or other medical device authorities. Any application of this model in healthcare or biomedical settings must comply with relevant regulatory requirements and undergo independent validation. Users assume full responsibility for how they use this model and any resulting consequences. The authors, contributors, and distributors disclaim any liability for damages, direct or indirect, resulting from model use. Users are responsible for ensuring compliance with data protection regulations (e.g., GDPR, HIPAA) when using it in research that involves patient data.
Log in or Sign Up to review the conditions and access this model content.
CytoSyn: a REPA-E Histopathology Image Generation Model
CytoSyn is a REPA-E [1] diffusion model trained on ~40M tiles, extracted from ~10k TCGA Diagnostic slides, achieving high-quality histopathology image generation at 224×224 resolution.
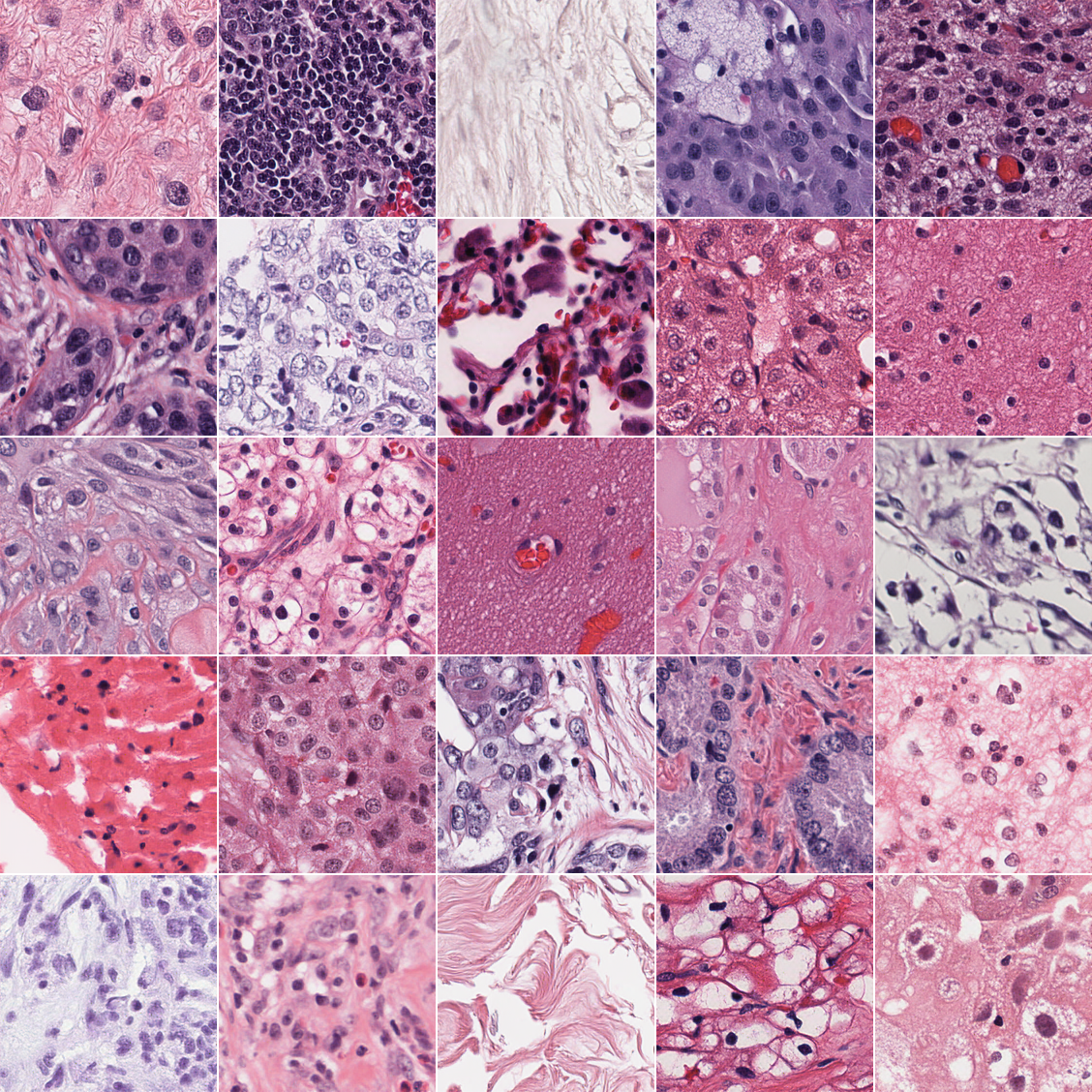
Figure 1: A sample of tiles sampled unconditionally with CytoSyn
The model consists of the following components:
- VAE: SD-VAE f8d4 [2],
- Latent Diffusion Transformer: SiT-XL/2 [3],
- Conditioning: H0-mini [4], a ViT-B/14 distilled from H-optimus-O [5], by Owkin & Bioptimus,
- Scheduler: Euler-Maruyama SDE scheduler [3].
Load REPA-E Model
from diffusers import DiffusionPipeline
import torch
device = torch.device('cuda')
pipeline = DiffusionPipeline.from_pretrained(
"Owkin-Bioptimus/CytoSyn",
custom_pipeline="Owkin-Bioptimus/CytoSyn",
trust_remote_code=True,
)
pipeline.to(device)
Load H0-mini for Conditioning
⚠️ CytoSyn uses H0-mini [CLS]-token for conditional generation. It must be loaded externally and the features must be extracted beforehand.
Note: You can get access to H0-mini on HuggingFace here.
import timm
from timm.data import resolve_data_config
from timm.data.transforms_factory import create_transform
# Load h0_mini encoder
h0_mini = timm.create_model(
"hf-hub:bioptimus/H0-mini",
pretrained=True,
mlp_layer=timm.layers.SwiGLUPacked,
act_layer=torch.nn.SiLU,
)
h0_mini = h0_mini.to(device)
h0_mini.eval()
# Get preprocessing transform
transform = create_transform(**resolve_data_config(h0_mini.pretrained_cfg, model=h0_mini))
Unconditional Generation
Generate histopathology images without conditioning:
# Generate 4 samples
output = pipeline(
num_images_per_prompt=4,
num_inference_steps=250,
guidance_scale=1.0, # No guidance for unconditional
)
images = output["images"]
# Save images
for i, img in enumerate(images):
img.save(f"sample_{i}.png")
Conditional Generation
Generate images conditioned on reference histopathology images:
from PIL import Image
# Load and preprocess conditioning image
conditioning_image = Image.open("reference.png").resize((224, 224))
img_tensor = transform(conditioning_image).unsqueeze(0).to(device)
# Extract h0_mini features (CLS token)
with torch.inference_mode():
h0_mini_embeds = h0_mini(img_tensor)[:, 0] # [1, 768]
# Generate conditioned samples with classifier-free guidance
output = pipeline(
h0_mini_embeds=h0_mini_embeds,
num_images_per_prompt=4,
num_inference_steps=250,
guidance_scale=2.5,
guidance_low=0.0,
guidance_high=0.75,
)
images = output["images"]
Software Dependencies
- torch>=2.0.0
- diffusers>=0.35.1
- timm>=0.9.0
- pillow
- huggingface-hub
Citation
Preprint coming soon.
In the meantime you can find more information about the model and its performance in this blog article.
Acknowledgements
Computing Resources
This work was granted access to the High-Performance Computing (HPC) resources of Meluxina, from LuxProvide, as part of a Euro-HPC grant under the allocation EHPC-AI-2024A04-020.
Code
CytoSyn was built using the REPA-E repository (MIT License).
License
The model is only available to academic and research institutions, for non-commercial use.
Contact
For questions, comments and issues, contact Thomas Duboudin ([email protected]).
References
Leng, X., Singh, J., Hou, Y., Xing, Z., Xie, S., & Zheng, L. (2025). Repa-e: Unlocking vae for end-to-end tuning with latent diffusion transformers. arXiv preprint arXiv:2504.10483.
Rombach, R., Blattmann, A., Lorenz, D., Esser, P., & Ommer, B. (2022). High-resolution image synthesis with latent diffusion models. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 10684-10695). arXiv:2112.10752
Ma, N., Goldstein, M., Albergo, M. S., Boffi, N. M., Vanden-Eijnden, E., & Xie, S. (2024, September). Sit: Exploring flow and diffusion-based generative models with scalable interpolant transformers. In European Conference on Computer Vision (pp. 23-40). Cham: Springer Nature Switzerland. arXiv:2401.08740
Filiot, A., Dop, N., Tchita, O., Riou, A., Dubois, R., Peeters, T., ... & Olivier, A. (2025, September). Distilling foundation models for robust and efficient models in digital pathology. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 162-172). Cham: Springer Nature Switzerland. arXiv:2501.16239 | HuggingFace
Saillard, C. and Jenatton, R. and Llinares-López, F. and Mariet, Z. and Cahané, D. and Durand, E. and Vert, J.P. (2024). H-optimus-0. URL: https://github.com/bioptimus/releases/tree/main/models/h-optimus/v0 | HuggingFace
- Downloads last month
- 59